the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Genetic variant of SPARC gene and its association with growth traits in Chinese cattle
Danyang Zhang
Jiawei Xu
Peng Yang
Yifan Wen
Hua He
Jiaxiao Li
Juntong Liang
Yining Zheng
Zijing Zhang
Xianwei Wang
Xiang Yu
Eryao Wang
Chuzhao Lei
Hong Chen
Yongzhen Huang
SPARC is a cysteine-rich acidic secreted protein. It is a non-collagen component of bone, which is widely distributed in humans and animals and plays an important role. SPARC has been found in a variety of human cancers (breast cancer, stomach cancer, ovarian cancer, etc.) and diabetes-related research. Especially the muscle and fat metabolism are closely related. In this study, we used a DNA pool to detect a new SNP site (g.12454T > C). A total of 616 samples of four breeds of Qinchuan cattle (QC, n=176), Xianan cattle (XN, n=160), Pinan cattle (PN, n=136) and Jiaxian cattle (JX, n=144) were analyzed and identified with ARMS-PCR. In addition, we correlated SNP with growth traits and showed significant correlation with growth traits such as rump length, hip width, and body length (p<0.05). Moreover, we tested the SPARC gene expression level in different tissues belonging to XN adult cattle (n=3) and found its high expression in muscle tissues (relative to the kidney). Further, we found the SNP is able to increase the SPARC expression level in skeletal muscle (n=12). According to statistical data, this SNP site may be applied to a molecular marker of an early marker-assisted selection for early growth of beef cattle.
The single nucleotide polymorphism (SNP) is the one nucleotide variation in the DNA sequence which will produce many polymorphisms in the genome of animals. It involves the transformation or transversion of a single nucleotide, which occurs in the sequence of the encoded protein or in the sequence of introns and intergenic regions (Dang et al., 2014). The SNP which happens in the CDS region will change and affect the protein if the mutation is the missense type. The other situation is the SNP happens in the CDS region, but it does not revise the coding of genes, which we call the synonymous mutation. The reason is the degeneracy of the codon. However, whatever the type of SNP, it will cause an impact of biological processes and sometimes it will cause phenotypic variation (Moravčíková et al., 2018; Carignano et al., 2018; Nakajima et al., 2018).
There are many methods of SNP genotype classification, such as RFLP, introduction mutation and direct sequencing. Moreover, the tetra-primer application refractory mutation system PCR (T-ARMS-PCR) is used as a low-cost, rapid genotyping assay (Hamajima et al., 2000).
The SPARC (secreted protein acidic and rich in cysteine) gene, which was first extracted by Termine et al. (1981), is a cysteine-rich acidic secreted protein, which is called the osteonectin or basement membrane 40 protein (Workman and Bradshaw, 2007). According to many current studies, the expression of SPARC protein is closely related to the occurrence of cancer (Koblinski et al., 2005; Mccabe et al., 2011; Alachkar et al., 2014). The SPARC protein can affect cell cycles, and it inhibits the cell proliferation by EF-hand to binding Ca2+ to influence the DNA synthesis (Sage et al., 1995).
SPARC is secreted by the adipocyte, which as a regulator in the extra cellular matrix inhibits fat formation and promotes fibrosis of adipose tissue, which is resistant to insulin. Recently, SPARC has become an important target molecule in the study of diseases such as obesity and diabetes (Harries et al., 2013). According to the research by selecting calf muscle tissue and using RT-PCR to detect the expression of the SPARC gene in muscle tissue, it was found that SPARC in db/db mouse muscle was highly expressed, suggesting that it may be related to muscle metabolism (Song et al., 2016). Due to its high amino acid homology, SPARC has similar functions in many animals. Therefore, its research on animals is of great significance. In kazak sheep and Tibetan sheep, such as the tail of the adipose tissue of the gene identification, SPARC expression may be associated with fat deposition. Sheep tails of excessive fat deposition will increase the investment and the cost of feed, thereby reducing the economic benefits of production (Guo et al., 2018).
SPARC is a kind of acid secreted protein that is rich in cysteine and widely distributed in the body. Because of its special structure of the area, it plays a role in the inhibition of cell cycle, cell adhesion and metastasis, and it has an effect on adjustment between cells and the matrix. At the same time, SPARC also can carry on the mediation to the tissue repair, angiogenesis and other functions (Alkabie et al., 2016). In this research, we found a SNP happens in SPARC gene in Chinese cattle breeds. The polymorphism of the mutation was tested in a large group of different Chinese cattle breeds, and the association was analyzed with the growth traits, which would be of benefit to cattle breeding and genetic research.
2.1 Experimental animals
The animals used in this study were four adult (24 mouth old) female breeds from China: Qinchuan cattle (QC, n=176, Fufeng, Shaanxi Province, China), Pinan cattle (PN, n=136, Xinye County, Henan Province, China), Xianan cattle (XN, n=160, Biyang County, Henan Province, China) and Jiaxian Red cattle (JX, n=144; Jia County, Pingdingshan City, Henan Province). Animal care and study protocols were in accordance with the Animal Care Commission of the College of Veterinary Medicine, Northwest A&F University. Then, the cattle samples were collected from the same breeding farm and the same batch. The body size data of growth traits are accurately measured and recorded according to the same criteria. They are in a state of uniform feeding, and there is no blood relationship between the three generations. And all breeds were feed under the same conditions.
2.2 Collection of blood samples and genomic DNA extraction
The samples obtained from the blood belong to each individual. Genomic DNA extracted by phenol chloroform method. The concentration and purity of the genomic DNA were tested and diluted to a uniform final concentration of 25 ng µL−1 for subsequent amplification assays.
2.3 Collection of different cattle tissues and cDNA extraction
The expression test sample is from XN adult female cattle (n=3). Samples included heart, liver, kidney, lung and muscle tissues. In order to research the relationship between SNP and gene transcription expression level, muscle tissues (n=12) were collected. The total RNA of all tissues was determined using the Trizol method of extraction following the manufacturer's instructions. The cDNA which we collected via RNA was reverse-transcribed using PrimeScript™ RT Reagent Kit with gDNA Eraser (Clontech, TaKaRa).
2.4 Primer design and amplification assay
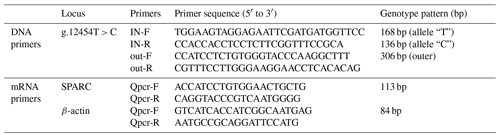
SNP primers were designed using the ARMS-primer method (http://primer1.soton.ac.uk/primer1.html, last access: 3 March 2019) based on the DNA sequence of the SPARC gene searched in the NCBI (NC_037334.1) and linked to the re-sequencing results. And the RNA expression test primers were designed based on SPARC gene mRNA sequence (NM_174464.2). All primer sequences are shown in Table 1.
The same amount of genomic DNA from 50 different individuals of cattle was mixed into a DNA pool for amplification of the gene of interest, and the volume of PCR reaction was 10 µL: the reaction system included 25 ng of genomic DNA (25 ng µL−1), 0.5 µL of internal and external primers (10 pmol µL−1), 5 µL 2 × Taq PCR Master Mix (GeneStar, Beijing, China), and 2 µL ddH2O. The procedure is as follows: the reactants are held at 95 ∘C for 5 min, at 94 ∘C for 30 s, down to 60 ∘C for 30 s, up to 72 ∘C for 25 s, the above steps for 40 cycles, and finally at 72 ∘C for 10 min. The PCR product is then sequenced to find mutations. The determination is typically made by sequencing the product in both forward and reverse directions (Sangon, Shanghai, China).
2.5 qPCR test of SPARC expression level
Fluorescence quantitative detection of SPARC expression levels in different tissues was performed using a CFX 96TM real-time quantitative RCR instrument (Bio-Rad, Hercules, CA, USA). The beta-actin gene stably expressed in bovine tissues was corrected as an internal reference.
2.6 SNP verification and statistical analysis
The SNP phenotype of SPARC gene was tested via the T-ARMS-PCR method, and the phenotype showed in 3.0 % agarose gel. The genotype frequency and allele frequency of SPARC gene SNPs were statistically analyzed using Excel software. In addition, SPSS V19.0 was used for the correlation analysis of SNP with various growth traits. According to the experimental design, a simplified model processing analysis was performed: Yijk = , where Yijk is the individual phenotypic record, μ the population mean, Tj the genotype effect, and Eijk the random error. The SPARC gene expression level was quantified by using an optimized method of comparing Ct (ΔΔCt) values (commonly referred to as 2−ΔΔCt). All samples were guaranteed at least three technical replicates, and the intensity ratio ± SD average was obtained therefrom.
3.1 SNP detection and gene phenotype analysis
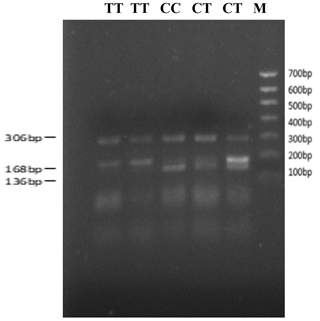
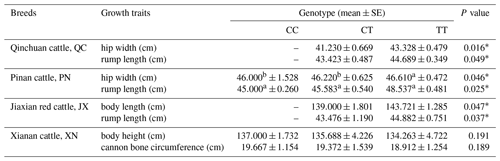
The bovine SPARC gene was mapped to chromosome 7, and the SNP (g.12454T > C, fourth exon) of the SPARC gene was found in the Chinese cattle genome (Fig. 1). The SNP is a synonymous mutation (59Ala > Ala). It is tested by PCR amplification using the ARMS-primer method. Thus, three genotypes were found at the SNP site (g.12454T > C). The length of target band showed in the gel of TT genotype was run into 306 and 168 bp, the length of band of CT genotype was 306, 168 and 136 bp, and the CC genotypic band was 306 and 136 bp (Fig. 2).
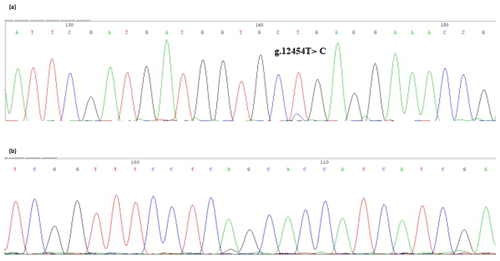
Figure 1Sequencing of the SNP of cattle SPARC gene. Note that the sequence test is the SPARC (exon 4) in Chinese cattle DNA pool. (a) Forward sequencing; (b) reverse sequencing.
3.2 Frequency statistics and PIC analysis
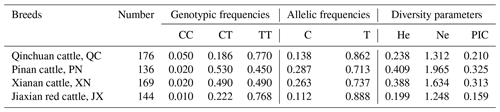
Statistical analysis of 616 cattle SPARC gene SNP typing found that in QC, PN, XN and JX, the frequency of allele T is much greater than the frequency of allele C. We further calculated He (gene heterozygosity), Ne (the number of effective alleles; the reciprocal of homozygotes), Ho (gene homozygous) and PIC (polymorphism information content) with the POPGENE software; the methods are as follows (Nei, 1973; Botstein et al., 1980):
The diversity parameter of PIC is 0.210, 0.325, 0.313 and 0.159 in QC, PN, XN and JX cattle, respectively. The PIC value of QC and JX < 0.25 indicates low genetic diversity. The PIC values of PN and XN > 0.25 and <0.5 indicate intermediate genetic diversity (Table 2).
3.3 The SPARC gene transcription level test
The result showed the transcription level of SPARC in five different tissues of adult bovine (Fig. 3). It was expressed in all tissues which we tested (heart, kidney, liver, muscle and lung), and the result showed differences in the tissue expression of the SPARC gene in adult cattle. The SPARC gene was most highly expressed in muscle tissues compared to others (relative to the kidney). On the contrary, it was most lowly expressed in the kidney compared other tissues.
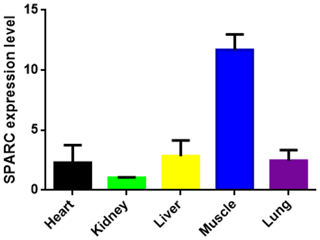
Figure 3The SPARC mRNA expression level in adult different tissues (relative to the kidney). Note the expression profiling of SPARC gene in different tissues in XN cattle. The values are the averages of three independent experiments measured by 2−ΔΔCt. Error bars represent the standard deviation (SD) (n=3), and the relative mRNA expression levels of SPARC gene are normalized; β-actin was used as an internal reference.
3.4 Correlation analysis of SPARC gene SNP and mRNA expression levels
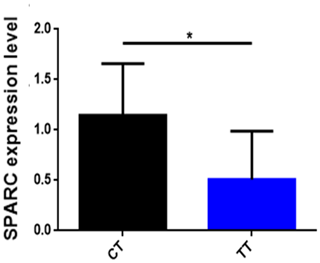
To find out whether the influence of SNP affects the mRNA expression level of the SPARC gene, we analyzed the correlation of SPARC SNP with mRNA expression levels in skeletal muscles from 12 adult cattle (Fig. 4). The results are shown in Fig. 4: the CT gene phenotype (n=6) has a high expression level of the SPARC gene compared to the TT gene phenotype (n=6) (p<0.05).
3.5 The association result between SNP of SPARC gene and phenotypic data
The results show that the SNP in QC and PN cattle has a significant influence on the rump length and hip width (P<0.05); moreover, the mutation also had a significant influence on the rump length and body length of JX cattle (P<0.05). In XN cattle, the SNP has a trend to affect the traits regarding the body height and cannon bone circumference. It shows an effect of CC type > CT type > TT type but not significantly (P>0.05). To summarize, these data results found that the SNP had a positive influence on the growth of skeletal muscle in the hindquarters of cattle, such as body length, hip length and hip width (Table 3).
Secreted protein acidic and rich in cysteine (SPARC), also known as osteonectin or BM-40, is the prototypical matricellular protein, which is secreted by many cells and distributed widely in the body, mainly distributed in the bone, cartilage and eye tissue (Scavelli et al., 2015). The main source of cells in the subcutaneous blood circulation is subcutaneous fat cells, which is a 32 kDa extra cellular matrix glycoprotein. The SPARC gene encodes a protein encoded by 298–304 amino acids. The protein has three structurally and functionally distinct modules. One is the amino acid terminal acidic calcium ion-binding region, the I region, which binds to copper ions homologous to follicle-binding elements. The second region is region II, and the third is the extra cellular calcium ion-binding region, region III. Based on such a binding region, it has an antigenic decision, inhibiting endothelial cell proliferation and angiogenesis. It also has functions such as decellularization (Salvatierra et al., 2015; Wong and Sukkar, 2016).
However, there are few reports on the genes of animals, especially cattle. According to our study, a SNP site of loci in SPARC gene is significantly associated with cattle body size. It was found to be significantly associated with character reflecting the development of the hindquarters of the cattle. The polymorphism statistics showed that the polymorphism of the mutation in the hybrid cattle of XN cattle and PN cattle was more polymorphic than that of the Chinese local cattle breed, which may be related to hybridization, and the mutation is suitable for molecular markers of yellow cattle breeding. The SPARC gene was highly expressed in skeletal muscle tissue. Although this mutation occurs on the fourth exon but belongs to a synonymous mutation (59Ala > Ala), it does not alter the protein coding. However, studies have also reported that synonymous mutations have a protein regulation function effect on the phenotype, etc., through other complex methods (Nackley et al., 2006; Kimchi-Sarfaty et al., 2007; Sauna and Kimchi-Sarfaty, 2011). And we also find that the SNP of the SPARC gene may change its expression in skeletal muscle tissue. Therefore, it is necessary to further study how the occurrence of this mutation affects the growth traits of cattle through more in-depth individual cell tests.
In this study, a SNP locus of SPARC gene was obtained by analyzing the mutation site of the SPARC gene. Among the four varieties tested, the traits of Qinchuan cattle, Pinan cattle and Jiaxian red cattle and the mutation of SPARC gene have a significant relationship (p<0.05). This mutation is of positive significance for early selection and breeding, and it provides new research directions and ideas.
The original data are available upon request to the corresponding author.
DZ and JX wrote the original draft. PY, YW, HH, JiL, JuL and YZ reviewed and edited the manuscript. ZZ, XW, XY and EW provided the technical and material. CL, HC and YH directed and supervised the project.
The authors declare that they have no conflict of interest.
This research has been supported by the Program of National Natural Science Foundation of China (grant no. 31601926), the National Beef Cattle and Yak Industrial Technology System (grant no. CARS-37), the Key Research & Development Plan of Shaanxi Province of China (General Project) (grant no. 2017NY-071), and the Key Science and Technology Program of Henan Province (grant nos. 152102110108 and 172102110062).
This paper was edited by Steffen Maak and reviewed by two anonymous referees.
Alachkar, H., Santhanam, R., Maharry, K., Metzeler, K. H., Huang, X. M., Kohlschmidt, J., Mendler, J. H., Benito, J. M., Hickey, C., Neviani, P., Dorrance, A. M., Anghelina, M., Khalife, J., Tarighat, S. S., Volinia, S., Whitman, S. P., Paschka, P., Hoellerbauer, P., Wu, Y. Z., Han, L., Bolon, B. N., Blum, W., Mrózek, K., Carroll, A. J., Perrotti, D., Andreeff, M., Caligiuri, M. A., Konopleva, A., Garzon, R., Bloomfield, C. D., and Marcucci G.: SPARC promotes leukemic cell growth and predicts acute myeloid leukemia outcome, J. Clin. Invest., 124, 1512–1524, https://doi.org/10.1172/JCI70921, 2014.
Alkabie, S., Basivireddy, J., Zhou, L. X., Roskams, J., Rieckmann, P., and Quandt, J. A.: SPARC expression by cerebral microvascular endothelial cells in vitro and its influence on blood-brain barrier properties, J. Neuroinflamm., 13, 225, https://doi.org/10.1186/s12974-016-0657-9, 2016.
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W.: Construction of a genetic linkage map in man using restriction fragment length polymorphisms, Am. J. Hum. Genet., 32, 314–331, https://doi.org/10.1016/0165-1161(81)90274-0, 1980.
Carignano, H. A., Roldan, D. L., Beribe, M. J., Raschia, M. A., Amadio, A., Nani, J. P., Gutierrez, G., Alvarez, I., Trono, K., Poil, M. A., and Miretti, M. M.: Genome-wide scan for commons snps affecting bovine leukemia virus infection level in dairy cattle, BMC Genomics, 19, 142, https://doi.org/10.1186/s12864-018-4523-2, 2018.
Dang, Y. L., Li, M. X., Yang, M. J., Cao, X. K., Lan, X. Y., Lei, C. Z., Zhang, C. L., Lin, Q., and Chen, H.: Identification of bovine NPC1 gene cSNPs and their effects on body size traits of Qinchuan cattle, Gene, 540, 153–160, https://doi.org/10.1016/j.gene.2014.03.001, 2014.
Guo, J. Z., Tao, H. X., Li, P. F., Li, L., Zhong, T., Wang, L. J., Ma, J. Y., Chen, X. Y., Song, T. Z., and Zhang, H. P.: Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds, Sci. Rep.-UK, 8, 10405, https://doi.org/10.1038/s41598-018-28719-w, 2018.
Hamajima, N., Saito, T., Matsuo, K., Kozaki, K., Takahashi, T., and Tajima, K.: Polymerase chain reaction with confronting two-pair primers for polymorphism genotyping, Cancer Sci., 91, 865–868, https://doi.org/10.1111/j.1349-7006.2000.tb01026.x, 2000.
Harries, L. W., McCulloch, L. J., Holley, J. E., Rawling, T. J., Welters, H. J., and Kos, K.: A role for SPARC in the moderation of human insulin secretion, PloS one, 8, e68253, https://doi.org/10.1371/journal.pone.0068253, 2013.
Kimchi-Sarfaty, C., Oh, J. M., Kim, I. W., Sauna, Z. E., Calcagno, A. M., Ambudkar, S. V., and Gottesman, M. M.: A “silent” polymorphism in the MDR1 gene changes substrate specificity, Science, 315, 525–528, https://doi.org/10.1126/science.1135308, 2007.
Koblinski, J. E., Kaplansinger, B. R., Vanosdol, S. J., Wu, M., Engbring, J. A., Wang, S., Goldsmith, C. M., Piper, J. T., Vostal, J. G., Harms, J. F., Welch, D. R., and Kleinman, H. K.: Endogenous osteonectin/sparc/bm-40 expression inhibits mda-mb-231 breast cancer cell metastasis, Cancer Res., 65, 7370–7377, https://doi.org/10.1158/0008-5472.can-05-0807, 2005.
Mccabe, N. P., Kerr, B. A., Madajka, M., Vasanji, A., and Byzova, T. V.: Augmented osteolysis in sparc-deficient mice with bone-residing prostate cancer, Neoplasia, 13, 31–39, https://doi.org/10.1593/neo.10998, 2011.
Moravčíková, N., Trakovická, A., Navrátilová, A., and Nádaský, R.: Associations between snps in bovine estrogen receptor gene and production traits in holstein cattle, J. Microbiol. Biotechn., 4, 83–85, https://doi.org/10.15414/jmbfs.2015.4.special2.83-85, 2018.
Nackley, A. G., Shabalina, S. A., Tchivileva, I. E., Satterfield, K., Korchynskyi, O., Makarov, S. S., Maixner, W., and Diatchenko, L.: Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure, Science, 314, 1930–1933, https://doi.org/10.1126/science.1131262, 2006.
Nakajima, A., Kawaguchi, F., Uemoto, Y., Fukushima, M., Yoshida, E., Iwamoto, E., Akiyama, T., Kohama, N., Kobayashi, E., Honda, T., Oyama, K., Mannen, H., and Sasazaki, S.: A genome-wide association study for fat-related traits computed by image analysis in japanese black cattle, Anim. Sci. J., 89, 743–751, https://doi.org/10.1111/asj.12987, 2018.
Nei, M.: Analysis of gene diversity in subdivided populations, P. Natl. Acad. Sci. USA, 70, 3321–3323, https://doi.org/10.1073/pnas.70.12.3321, 1973.
Sage, E. H., Bassuk, J. A., Yost, J. C., Folkman, M. J., and Lane T. F.: Inhibition of endothelial cell proliferation by SPARC is mediated through a Ca2+-binding ef-hand sequence, J. Cell. Biochem., 57, 127–140, https://doi.org/10.1002/jcb.240570113, 1995.
Salvatierra, E., Alvarez, M. J., Leishman, C. C., Baquero, E. R., Lutzky, V. P., Chuluyan, H. E., and Podhajcer, O. L.: SPARC Controls Melanoma Cell Plasticity through Rac1, PloS one, 10, e0134714, https://doi.org/10.1371/journal.pone.0134714, 2015.
Sauna, Z. E. and Kimchi-Sarfaty, C.: Understanding the contribution of synonymous mutations to human disease, Nat. Rev. Genet., 12, 683–691, https://doi.org/10.1038/nrg3051, 2011.
Scavelli, K., Chatterjee, A., and Rhee, D. J.: Secreted Protein Acidic and Rich in Cysteine in Ocular Tissue, J. Ocul. Pharmacol. Th., 31, 396–405, https://doi.org/10.1089/jop.2015.0057, 2015.
Song, H. Y., Yang, X. Y., and Lei, D.: Increased SPARC expression in skeletal muscle and adipose tissue of db/db mice, Int. J. Clin. Exp. Patho., 9, 8274–8279, 2016.
Termine, J. D., Kleinman, H. K., Whitson, S. W., Conn, K. M., McGarvey, M. L., and Martin, G. R.: Osteonectin, a bone-specific protein linking mineral to collagen, Cell, 26, 99–105, https://doi.org/10.1016/0092-8674(81)90037-4, 1981.
Wong, S. L. and Sukkar, M. B.: The SPARC protein: an overview of its role in lung cancer and pulmonary fibrosis and its potential role in chronic airways disease, Brit. J. Pharmacol., 174, 3–14, https://doi.org/10.1111/bph.13653, 2016.
Workman, G. and Bradshaw, A. D.: Production and purification of recombinant human SPARC, Method Cell. Biol., 143, 335, https://doi.org/10.1016/bs.mcb.2017.08.020, 2017.