the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Polymorphisms in pattern recognition receptor genes of indigenous and White Leghorn breeds of chicken
Santosh Haunshi
Arun Kumar Burramsetty
Kannaki Ramasamy
Rudra Nath Chatterjee
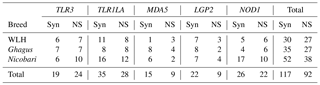
Functional polymorphisms in pattern recognition receptors (PRRs) modulate innate immunity and play a crucial role in resistance or susceptibility to diseases. The present study was carried out to explore polymorphic patterns in the coding sequences of PRR genes TLR3, TLR1LA (TLRs), MDA5, LGP2 (RLRs) and NOD1 (NLR) in chicken breeds of India, namely Ghagus (GH), Nicobari (NB) and the exotic White Leghorn (WLH) breed. Out of 209 SNPs observed in five genes among three breeds, 117 were synonymous (Syn) and 92 were non-synonymous (NS) SNPs. In TLR genes the highest polymorphism was observed in NB (16, 28) compared to GH (14, 16) and WLH (13, 19) breeds. In the MDA5 gene the highest polymorphism was observed in GH (12) compared to NB (eight) and WLH (four) breeds. However, an almost similar level of polymorphism was observed in the LGP2 gene among the three breeds. In the NOD1 gene, the highest polymorphism was observed in NB (27), followed by WLH (11) and GH (10) breeds. The overall highest number of SNPs was observed in NB (90), followed by GH (62) and the WLH (57) breed. With regard to variation in polymorphism among different classes of PRRs, the study revealed the highest polymorphism in TLRs compared to NOD1 and the RLR class of PRRs. Further, the domain locations of various Syn and NS SNPs in each PRR among the three breeds were identified. In silico analysis of NS SNPs revealed that most of them had a neutral effect on protein function. However, two each in TLR1LA and LGP2 and one in the MDA5 gene were predicted to be deleterious to protein function. The present study unravelled extensive polymorphism in the coding sequences of the TLR and NLR class of PRR genes, and the polymorphism was higher in indigenous chicken breeds.
- Article
(186 KB) -
Supplement
(213 KB) - BibTeX
- EndNote
Pattern recognition receptors (PRRs) are highly conserved germ-line-encoded molecules which recognize pathogen-associated molecular patterns (PAMPs) that are an integral part of pathogens. Recognition of PAMPs by PRRs leads to activation of innate immune response by inducing type I interferons and pro-inflammatory cytokines. Thus, PRRs work as first line of defence against invading pathogens. In chickens, three classes of PRRs, such as Toll-like receptors (TLRs), retinoic acid-inducible gene-I-like (RIG-I) receptors (RLRs), and nucleotide-binding and oligomerization-domain-like receptors (NLRs), have been described (Chen et al., 2013). The role of TLRs in the innate immune system and resistance to various infectious diseases has been well established (Keestra et al., 2013). A previous study reported the association of polymorphism in the TLR4 gene with susceptibility to Salmonella enterica serovar Typhimurium infection in chickens (Leveque et al., 2003). Recently, the importance of other classes of PRRs such as RLR (Hayashi et al., 2014) and NLRs (Tao et al., 2015, 2017) in various infectious diseases was reported. Genetic variation in the coding sequences (CDSs) of PRRs, which results in changes in amino acid sequence, has the potential to affect the function of protein receptors and thereby modulate the resistance or susceptibility to infectious diseases (Jaeger et al., 2015). Therefore, SNPs in the CDSs of PRR genes may alter the activity of PRRs and thereby affect the resistance or susceptibility to infectious diseases. This is one of the mechanisms by which the genetic make-up of an organism can influence resistance and tolerance to infectious agents. Indigenous chicken breeds are known for their hardiness and ability to survive under adverse conditions with little or no healthcare. Natural selection over the years under resource-poor suboptimum conditions made indigenous chickens more robust and tolerant or resistant to various diseases, particularly of bacterial and parasitic origin. On the other hand, exotic breeds such as the White Leghorn have been subjected to genetic improvement for economic traits and may not be that hardy or resistant to diseases (Besbes, 2009). Therefore, the present study was carried out with the aim of exploring and comparing polymorphic patterns in PRR genes such as TLR3, TLR1LA, MDA5, LGP2 and NOD1 in chicken breeds of India (Ghagus and Nicobari) vis a vis the exotic White Leghorn breed.
2.1 Experimental birds
Three breeds of chicken, namely Ghagus (GH), Nicobari (NB) and White Leghorn (WLH), were used in this study. GH and NB are indigenous to India: GH is from mainland India (Karnataka), while NB is from the Nicobar Islands. Pedigreed random mating populations of these breeds are maintained at the institute farm of the ICAR Directorate of Poultry Research, Hyderabad. Representative birds of the same age (four males) from each breed were selected and sacrificed for collection of spleen samples. Samples were collected on ice and processed immediately for RNA isolation. The experiment was carried out with the approval of the Institutional Animal Ethics Committee (IAEC).
2.2 RNA extraction and cDNA preparation
Total RNA from spleen samples was extracted using TRIzol (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. RNA thus obtained was quantified spectrophotometrically and converted to cDNA using a ProtoScript II First Strand cDNA conversion kit (New England Biolab Inc., MA, USA) following the manufacturer's instructions. Briefly, a reaction volume of 20 µL containing 1 µg of RNA, 5 µM oligo d(T)23 VN primer, 10 µL of reaction mix (2×) and 2 µL of enzyme mix (10×) was incubated at 42 ∘C for 1 h to convert RNA into cDNA, and the reaction was inactivated at 80 ∘C for 5 min. The converted cDNA was confirmed by amplifying a 150 bp fragment of the β-actin gene, and positive samples were stored at −20 ∘C until further use as a template for PCR reactions.
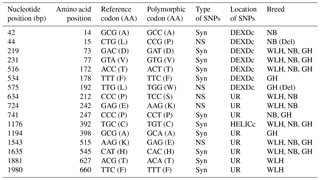
Table 2Polymorphisms in the TLR3 gene of different chicken breeds.
LRR: leucine-rich repeat domain, LRR_CT: leucine-rich repeat C-terminal domain, TIR: the Toll–interleukin-1 receptor homology domain, NS: non-synonymous, Syn: synonymous, UR: unknown region, GH: Ghagus, NB: Nicobari, WLH: White Leghorn.
2.3 Design of primers
Primer pairs for amplifying and sequencing the coding regions of PRR genes (TLR3, TLR1LA, LGP2, MDA5 and NOD1) were designed using the online primer designing tool Primer3Plus (http://www.bioinformatics.nl/primer3plus, last access: 12 March 2016) with sequences retrieved from the UCSC genome browser. Primers were checked for their specificity with Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast, last access: 4 May 2016) and used for PCR amplification and sequencing (Table S1 in the Supplement).
2.4 PCR amplification and sequencing
Fragments of coding regions were amplified through PCR. The reaction was carried out in a 20 µL volume containing 4 µL of 5× high-fidelity buffer, 0.5 U of KAPA HiFi HotStart DNA polymerase (Kapa Biosystems, Cape Town, South Africa), 400 µM dNTPs, 0.2 µM each of forward and reverse primers, and 2 µL of cDNA template. PCR amplification was carried out using initial denaturation at 95 ∘C for 5 min, followed by 40 cycles at 98 ∘C for 20 s, annealing at Ta (Table S1) for 30 s and extension at 72 ∘C for 40 s, with a final extension at 72 ∘C for 5 min. Amplified products were separated on agarose gel and desired fragments were gel eluted using a QIAquick gel extraction kit (Qiagen GmbH, Hilden, Germany) following the manufacturer's instructions. These eluted products were sequenced bidirectionally using BigDye terminator sequencing on an ABI 3730 sequencer (Bioserve Biotehnologies Pvt. Ltd., Hyderabad, India). A stepwise direct sequencing strategy was chosen using primer pairs designed to produce overlapping fragments. This way it was possible to obtain an unambiguous sequence covering the entire coding sequence of PRR genes.
The coding sequences of all five genes from the three breeds were deposited in GenBank. The accession numbers are as follows. TLR3: MF576160 (WLH), MF576161 (NB) and MF576162 (GH); TLR1LA: MF563596 (WLH), MF563597 (NB) and MF563598 (GH); MDA5: MF563590 (GH), MF563591 (WLH) and MF563592 (NB); LGP2: MF563593 (WLH), MF563594 (NB) and MF563595 (GH); NOD1: MF576163 (WLH), MF576164 (NB) and MF576165 (GH).
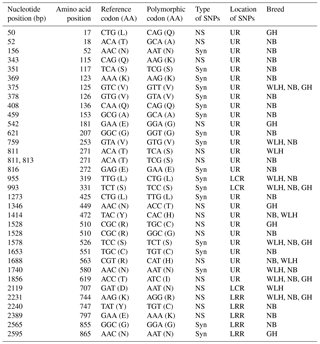
Table 3Polymorphisms in the TLR1LA gene of different chicken breeds.
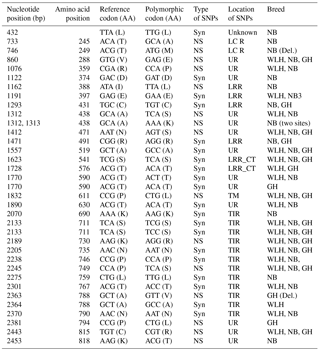
LRR: leucine-rich repeats, LRR_CT: leucine-rich repeats C-terminal domain NS: non-synonymous, Syn: synonymous, UR: unknown region, GH: Ghagus, NB: Nicobari, WLH: White Leghorn, Del.: deleterious, TIR: the Toll–interleukin-1 receptor homology domain.
2.5 Sequence analysis
Sequences in the forward and reverse directions of each fragment were aligned, checked for misread bases and edited using the CodonCode aligner v5.0.1 (CodonCode Corporation, USA). Thus, consensus sequences were multiple aligned with the ClustalW function in MEGA7 (https://www.megasoftware.net, last access: 11 April 2017) against respective reference sequences of red junglefowl (Gallus gallus). Synonymous (Syn) and non-synonymous (NS) SNPs were recorded and analysed for their effect (neutral or deleterious) of amino acid substitution on protein function using an online tool, the protein variation effect analyser (https://provean.jcvi.org/index.php, last access: 5 May 2017). Further, a simple modular architecture research tool (SMART) was used to identify the locations of SNPs on different domains of PRRs (Letunic et al., 2012).
Coding sequences of five genes belonging to three different classes of PRRs (TLRs, RLRs and NLRs) were studied for functional polymorphisms in GH, NB and WLH breeds. A total of 209 SNPs in five genes were observed among the three breeds: of these, 117 were Syn and 92 were NS SNPs. The highest polymorphism was observed in TLRs compared to NOD1 and the RLR class of PRRs. NS SNP results in the substitution of amino acid, and that leads to changes in the functions of receptors which can be advantageous, neutral or deleterious to the functions of proteins, while variation due to Syn substitution is expected to be neutral (Downing, et al., 2010). Amino acid substitutions due to polymorphism in the leucine-rich repeats (LRR) domain of PRRs may affect the ability of receptors to recognize PAMPs, while those in the transmembrane domain may lead to defects in intracellular receptor transport. Finally, those in the internal domain may result in altered interaction with adaptor proteins or disrupted dimerization (Kutikhin et al., 2013). Gene-wise SNPs in the CDSs of various PRR genes are described in the following sections.
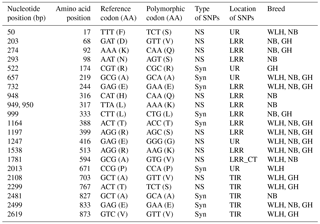
Table 4Polymorphisms in the MDA5 gene of different chicken breeds.
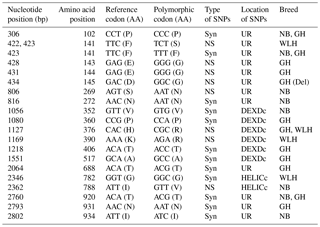
DEXDc: DEAD-like helicase superfamily, HELICc: helicase superfamily C-terminal domain, NS: non-synonymous, Syn: synonymous, UR: unknown region, TM: transmembrane, GH: Ghagus, NB: Nicobari, WLH: White Leghorn, Del: deleterious.
3.1 TLR3
TLR3 is an intracellular type I transmembrane receptor which recognizes the dsRNA of pathogens (viruses) and is known for its role in antiviral innate immunity in chickens. In this gene (CDS: 2691 bp) the highest number of NS SNPs was observed in NB compared to GH and WLH breeds (Table 1). A total of 10 NS SNPs were present in the extracellular domain, of which 8 were in the LRR domain, 1 was in the LRR_CT domain and 1 was in the unknown region, while only 2 NS SNPs were present in the intracellular TIR domain. Relatively higher polymorphism was observed in NB compared to the other two breeds. Polymorphism was higher at the LRR domain of TLR3 of NB, as 8 out of 10 NS SNPs observed were present in the extracellular LRR domain. Four out of 10 NS SNPs (A293G, T948A, T949A and T950A) were exclusive to NB and two NS SNPs (A203T and A274C) were shared between the NB and GH breeds, while no NS SNP was found to be exclusive to the WLH breed (Table 2). All these NS SNPs located in the LRR domain of TLR3 might play a role in the modulation of innate immunity since PAMPs are known to bind to the LRR domain of TLR3. Polymorphism in the coding sequence of the TLR3 gene was also reported previously in different chicken breeds (Ruan et al., 2015). Similar to the present findings, higher NS SNPs were reported in Chinese native chickens compared to the WLH breed. At least 10 NS SNPs (T50C, A203T, A274C, A293G, T999G, G1197C, G1538A, C1781T, C2108T and A2299T) were shared between chicken breeds in this study and Chinese breeds (Huang et al., 2012). Various studies have reported the relationship between polymorphisms in the TLR3 gene and resistance or susceptibility to different viral diseases in human beings (Kindberg et al., 2011; Sironi et al., 2012; Svensson et al., 2012; Lee et al., 2013; Studzińska et al., 2017). Similar relationships between polymorphisms in the TLR3 gene and resistance or susceptibility to various diseases could exist in chickens as well.
3.2 TLR1LA
The TLR1LA gene located on chromosome 4 of chickens is involved in innate immunity against bacterial infections. The TLR1LA receptor recognizes triacylated and diacylated lipopeptides as PAMPs through the leucine-rich repeat domain (Keestra et al., 2013). In this gene (CDS: 2188 bp partial) the highest number of NS SNPs was also seen in NB compared to GH and WLH breeds (Table 1). Out of 12 NS SNPs observed in NB, 6 (A733G, C746T, A1162T, G1312A, C1313A and A2453C) were exclusive to this breed, while only 2 (C2363T and C2381T) out of 8 NS SNPs observed were exclusive to the GH breed. No NS SNP was found to be exclusive to the WLH breed (Table 3). Five out of six exclusive NS SNPs of NB are located in the LRR or LCR domain of TRLA1LA where PAMPs are known to bind, and hence these NS SNPs might play a key role in innate immunity. Further, substitution of amino acids in the TIR domain (K730R, P749S and A788V) of TLR1LA may influence signal transduction as TIR is the functional domain for the subsequent recruitment of intracellular adapter proteins (Werling et al., 2009). Functional polymorphisms in human TLR1 and TLR2 genes were reported to be associated with resistance or susceptibility to various bacterial and fungal diseases (Rosentul et al., 2014; Jaeger et al., 2015). Extensive polymorphism in the TLR1LA gene was also reported in Chinese native and commercial chicken breeds (Ruan and Zheng, 2011), and at least 13 SNPs (Syn and NS) were shared between breeds in the present study and those studied by Ruan and Zheng (2011). Out of eight NS substitutions located in the extracellular domain of TLR1LA (Table 3), four substitutions, namely T245A, T249M, R359P and N471S, were common to indigenous chicken breeds of India and China. Six NS substitutions were located in the intracellular domain of TLR1LA, of which two NS substitutions (K730R and P749S) were shared with Chinese native breeds. One polymorphic site (P611L) located in the transmembrane domain among the three breeds was also reported in Chinese breeds (Ruan and Zheng, 2011).
3.3 MDA5
The MDA5 gene, also known as IFIH1 (CDS: 3006 bp), located on chromosome 7 codes for melanoma-differentiation-associated protein 5 (MDA5) and is an important member of the RLR class of PRRs. In this gene the highest number of NS SNPs was observed in GH, followed by the WLH and NB breed (Table 1). Most of the NS SNPs (five) were located in an unknown region, while two were located in DEXDc (GH and WLH) and one in the HELICc domain (NB) of the MDA5 receptor (Table 4). Out of four NS SNPs observed in GH, three (A428G, A431G and A434G) were exclusive to this breed; two NS SNPs (G806A and A2362G) are exclusive to NB and two NS SNPs (T422C and A1169G) are exclusive to WLH. Since most of the NS SNPs were located in an unknown region (domain) of the MDA5 receptor, their possible role in innate immunity could not be delineated (Table 4). MDA5 is one of the cytosolic PRRs that recognizes dsRNA and poly (I : C), a synthetic analogue. MDA5 was reported to be involved in the induction of chicken IFN β production against avian influenza virus infection (Hayashi et al., 2014). Further, it was reported that MDA5 recognizes the virus responsible for infectious bursal disease, and that leads to the activation of MDA5-related innate immunity and the upregulation of MHC class I (Lee et al., 2014). Polymorphism in the IFIH1–MDA5 gene was linked to spontaneous clearance of the hepatitis C virus infection in humans (Hoffmann et al., 2015). Further, a link indicating polymorphism in the MDA5 gene as a risk factor for the enterovirus 71 infection in humans was reported (Pang et al., 2014). However, the influence of polymorphism in the MDA5 gene on resistance or susceptibility to various viral diseases in chickens has yet to be reported.
3.4 LGP2
The laboratory of genetics and physiology 2 (LGP2–DHX58) gene (CDS: 2025 bp) is another cytosolic PRR belonging to the RLR class. LGP2, located on chromosome 27, encodes for the cytoplasmic DEx(D/H)-box helicase receptor that recognizes both ssRNA and dsRNA of virus origin (Zhu et al., 2014). An almost similar level of polymorphism was observed among the three breeds in the coding sequence of this gene. Only two NS SNPs were located on the DEXDc domain of the LGP2 receptor, while the remaining three were located on an unknown region (domain) of the LGP2 receptor (Table 5). One NS SNP (T44C) in Nicobari and one NS SNP (T575G) in GH were exclusive to these breeds and were located on the DEXDc domain of the LGP2 gene (Table 5). LGP2 is considered to be essential for the augmentation of MDA5-dependent signaling (Zhu et al., 2014) since it functions as a positive regulator of antiviral responses (Satoh et al., 2010). It is also reported to compensate for the lack of RIG-I PRR in chickens (Uchikawa et al., 2016). Polymorphism in the LGP2 gene in grass carp (Ctenopharyngodon idella) was reported to be associated with resistance or susceptibility to grass carp reovirus infection (Wan et al., 2013).
3.5 NOD1
The nucleotide-binding oligomerization-domain-containing protein 1 (NOD1) gene, located on chromosome 2, codes (CDS: 2856 bp) for an NLR-type intracellular PRR. This PRR is comprised of a caspase recruitment domain at the N terminus, seven leucine-rich repeat regions at the C terminus, and one NACHT domain between the N and C termini (Tao et al., 2015). In the NOD1 gene the highest number of NS SNPs was observed in NB compared to GH and WLH breeds (Table 1). Most of the NS SNPs were located in the unknown region of the protein, while three NS SNPs were present in the LRR domain and one in low-complexity region of the receptor (Table 6). Six NS SNPs (A52G, C343A, A811T, A813G, C1528G, A2240G and G2389A) were exclusive to NB and four NS SNPs (T50A, A542G, A1346C and C1528T) were exclusive to GH, while only two NS SNPs (A271T and G707A) were exclusive to WLH (Table 6). All exclusive SNPs of GH and the majority of exclusive SNPs of NB were situated in an unknown region (domain) of the NOD1, while two SNPs (A2240G and G2389A) of NB were located in the LRR domain of NOD1 that constitutes the recognition motif for PAMPs. All these NS SNPs might play a role in innate immunity as they can alter the structure or function of receptors by virtue of change in amino acid composition and sequence. NOD1 initiates innate immunity by recognition of a specific bacterial molecule: peptidoglycan as a ligand. Subsequent to ligand binding, the NOD1 receptor undergoes conformation changes and self-oligomerization and induces nuclear factor κB (NF κB) activation independent of TLRs, resulting in the production of pro-inflammatory cytokines, chemokines and antimicrobial peptides (Shaw et al., 2008). Expression of the NOD1 gene is found to be induced in Salmonella enterica serovar Enteritidis (Tao et al., 2015) and Salmonella pullorum (Tao et al., 2017) infections in chickens (Tao et al., 2015). Polymorphism in the NOD1 gene in humans was reported to be linked with Helicobacter pylori pathogenesis (Hofner et al., 2007; Kara et al., 2010), Chlamydia trachomatis infection (Branković et al., 2015) and cytomegalovirus control (Fan et al., 2016). The NS SNPs observed in this study could play a role in resistance to various infectious diseases in chickens, and their effect on innate immunity needs to be investigated in detail through pathogen challenge studies.
The study unravelled extensive single nucleotide polymorphisms in the coding sequences of PRR genes, and the polymorphism was higher in TLRs and NOD1 genes compared to RLR genes. With regards to polymorphism in the three breeds, the highest polymorphism was observed in indigenous chicken NB compared to GH and WLH breeds. The effects of functional polymorphisms in PRRs on resistance or susceptibility to various infectious diseases have been well established in humans and other domestic animals. Therefore, we speculate that polymorphisms in the CDSs of PRR genes could influence the resistance or susceptibility to various infectious diseases in chickens as well. To the best of our knowledge, few reports exist in the literature about the association of polymorphisms in PRR genes with disease resistance or susceptibility in chickens. Therefore, further studies are needed to examine the functional effects of these NS SNPs in PRR genes on resistance or susceptibility to various diseases in chickens. In silico analysis of NS SNPs for their possible effect on protein function revealed that most of them had a neutral effect on protein function. However, five SNPs (two each in TLR1LA and LGP2 and one in the MDA5 gene) were predicted to be deleterious in nature. These deleterious substitutions are of greater interest as they can affect the structure or function of the protein receptor.
The data from the paper are available upon request from the corresponding author.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-61-441-2018-supplement.
SH conceived and designed the study and drafted the paper. AKB conducted the experiment and analysed the sequences; TRK was involved in drafting the paper. RNC supported the study and critically reviewed the paper. All authors contributed to the discussion of the results and approved the final draft of the paper.
The authors declare that they have no conflict of interest.
The assistance of Konadaka Sri Rajaravindra, Farm Manager and Hatchery
Manager in carrying out this study is gratefully acknowledged. This study was
funded by the Science and Engineering Research Board (SERB) of the Ministry
of Science and Technology, Government of India (SB/SO/AS-015/2014).
Edited by: Steffen Maak
Reviewed by: two anonymous referees
Besbes, B.: Genotype evaluation and breeding of poultry for performance under sub-optimal village conditions, World's Poult. Sci. J., 65, 260–271, 2009.
Branković, I., van Ess, E. F., Noz, M. P., Wiericx, W. A., Spaargaren, J., Morré, S. A., and Ouburg, S.: NOD1 in contrast to NOD2 functional polymorphism influence Chlamydia trachomatis infection and the risk of tubal factor infertility, Pathog. Dis., 73, 1–9, 2015.
Chen, S., Cheng, A., and Wang, M.: Innate sensing of viruses by pattern recognition receptors in birds, Vet. Res., 44, 1–12, https://doi.org/10.1186/1297-9716-44-82, 2013.
Downing, T., Lloyd, A. T., O'Farrelly, C., and Bradley, D. G.: The differential evolutionary dynamics of avian cytokine and TLR gene classes, J. Immunol., 184, 6993–7000, 2010.
Fan, Y., Roy, S., Mukhopadhyay, R., Kapoor, A., Priya, D., Wojcik, G. L., Pass, R. F., and Arav-Boger, R.: Role of nucleotide-binding oligomerization domain 1 (NOD1) and its variants in human cytomegalovirus control in vitro and in vivo, P. Natl. Acad. Sci. USA, 113, E7818–E7827, 2016.
Hayashi, T., Watanabe, C., Suzuki, Y., Tanikawa, T., Uchida, Y., and Saito, T.: Chicken MDA5 senses short double-stranded RNA with implications for antiviral response against avian influenza viruses in chicken, J. Innate Immun., 6, 58–71, 2014.
Hoffmann, F., Schmidt, A., Chevillotte, M. D., Wisskirchen, C., Hellmuth, J. C., Willms, S., Gilmore, R. H., Glas, J., Folwaczny, M., Müller, T., Berg, T., Spengler, U., Fitzmaurice, K., Kelleher, D., Reisch, N., Rice, C. M., Endres, S., and Rothenfusser, S.: Polymorphisms in MDA-5 link protein function to clearance of hepatitis C virus, Hepatology, 61, 460–470, 2015.
Hofner, P., Gyulai, Z., Kiss, Z. F., Tiszai, A., Tiszlavicz. L., Tóth, G., Szõke, D., Molnár, B., Lonovics, J., Tulassay, Z., and Mándi, Y.: Genetic polymorphisms of NOD1 and IL-8, but not polymorphisms of TLR4 genes, are associated with Helicobacter pylori-induced duodenal ulcer and gastritis, Helicobactor, 12, 124–131, 2007.
Huang, Y. Q., Chen, W., Huang, Y. H., Shi, X. W., Deng, M., Kang, X. T., and Li, N.: Haplotypic variation and characteristics across the toll – like receptor 3 locus in chickens, Anim. Genet., 43, 343–347, 2012.
Jaeger, M., Stappers, M. H. T., Joosten, L. A. B., Gyssens, I. C., and Netea, M. G.: Genetic variation in pattern recognition receptors: functional consequences and susceptibility to infectious disease, Future Microbiol., 10, 989–1008, 2015.
Kara, B., Akkiz, H., Doran, F., Bayram, S., Erken, E., Gumurdullu, Y., and Sandikci, M.: The significance of E266K polymorphism in the NOD1 gene on Helicobacter pylori infection: an effective force on pathogenesis?, Clin. Exp. Med., 10, 107–112, 2010.
Keestra, A. M., de Zoete, M. R., Bouwman, L. I., Vaezirad, M. M., and Putten, J. P.: Unique features of chicken Toll-like receptors, Dev. Comp. Immunol., 41, 316–323, 2013.
Kindberg, E., Vene, S., Mickiene, A., Lundkvist, A., Lindquist, L., and Svensson, L.: A functional Toll-like receptor 3 gene (TLR3) may be a risk factor for tick-borne encephalitis virus (TBEV) infection, J. Infect. Dis., 203, 523–528, 2011.
Kutikhin, A. G. and Yuzhalin, A. E.: Pattern recognition receptors, gene polymorphisms, and cancer: A Double-edged sword In: Genomics of pattern recognition receptors: Applications in oncology and cardiovascular diseases, Springer Basel, 27–32, https://doi.org/10.1007/978-3-0348-0688-6, 2013.
Lee, C. C., Wu, C. C., and Lin, T. L.: Chicken melanoma differentiation-associated gene 5 (MDA5) recognizes infectious bursal disease virus infection and triggers MDA5-related innate immunity, Arch. Virol., 159, 1671–1686, 2014.
Lee, S. O., Brown, R. A., and Razonable, R. R.: Association between a functional polymorphism in Toll-like receptor 3 and chronic hepatitis C in liver transplant recipients, Transpl. Infect. Dis., 15, 111–119, 2013.
Letunic, I., Doerks, T., and Bork, P.: SMART 7: Recent updates to the protein domain annotation resource, Nucleic Acids Res., 40, D302–D305, 2012.
Leveque, G., Forgetta, V., Morroll, S., Smith, A. L., Bumstead, N., Barrow, P., Loredo-Osti, J. C., Morgan, K., and Malo, D.: Allelic variation in TLR4 is linked to susceptibility to Salmonella enterica serovar Typhimurium infection in chickens, Infect. Immun., 71, 1116–1124, 2003.
Pang, L., Gong, X., Liu. N., Xie, G., Gao, W., Kong, G., Li, X., Zhang, J., Jin, Y., and Duan, Z.: A polymorphism in melanoma differentiation-associated gene 5 may be a risk factor for enterovirus 71 infection, Clin. Microbiol. Infect., 20, O711—O717, 2014.
Rosentul, D. C., Delsing, C. E., Jaeger. M., Plantinga, T. S, Oosting, M., Costantini, I., Venselaar, H., Joosten, L. A., van der Meer, J. W., Dupont, B., Kullberg, B. J., Sobel J. D., and Netea, M. G.: Gene polymorphisms in pattern recognition receptors and susceptibility to idiopathic recurrent vulvovaginal candidiasis, Front. Microbiol., 5, 483, https://doi.org/10.3389/fmicb.2014.00483, 2014.
Ruan, W., An, J., and Wu, Y.: Polymorphisms of chicken TLR3 and 7 in different breeds, PLoS ONE, 10, e0119967, https://doi.org/10.1371/journal.pone.0119967, 2015.
Ruan, W. K. and Zheng, S. J.: Polymorphisms of chicken toll-like receptor 1 type 1 and type 2 in different breeds, Poult. Sci., 90, 1941–1947, 2011.
Satoh, T., Kato, H., Kumagai, Y., Yoneyama, M., Sato, S., Matsushita, K., Tsujimura, T., Fujita, T., Akira, S., and Takeuchi, O.: LGP2 is a positive regulator of RIG-I and MDA5 mediated antiviral responses, P. Natl. Acad. Sci. USA, 107, 1512–1517, 2010.
Shaw, M. H., Reimer, T., Kim, Y. G., and Nuñez, G.: NOD-like receptors (NLRs): bona fide intracellular microbial sensors, Curr. Opin. Immunol. 20, 377–382, 2008.
Sironi, M., Biasin, M., Cagliani, R., Forni, D., Luca, M. D., Saulle, I., Lo Caputo, S., Mazzotta, F., Macías, J., Pineda, J. A., Caruz, A., and Clerici, M..: A common polymorphism in TLR3 confers natural resistance to HIV-1 infection, J. Immunol., 188, 818–823, 2012.
Studzińska, M., Jabłońska, A., Wiśniewska-Ligier, M., Nowakowska, D., Gaj, Z., Leśnikowski, Z. J., Woźniakowska-Gęsicka, T., Wilczyński, J., and Paradowska, E.: Association of TLR3 L412F Polymorphism with Cytomegalovirus Infection in Children, PLoS One, 12, e0169420, https://doi.org/10.1371/journal.pone.0169420, 2017.
Svensson, A., Tunback, P., Nordstrom, I., Padyukov, L., and Eriksson, K.: Polymorphisms in Toll-like receptor 3 confer natural resistance to human herpes simplex virus type 2 infection, J. Gen. Virol., 93, 1717–1724, 2012.
Tao, Z. Y., Zhu, C. H., Shi, Z. H., Song, C., Xu, W. J., Song, W. T., Zou, J. M., and Qin, A. J.: Molecular characterization, expression, and functional analysis of NOD1 in Qingyuan partridge chicken, Genet. Mol. Res., 14, 2691–2701, 2015.
Tao, Z., Zhu, C., Song, W., Xu, W., Zhang, H., Liu, H., and Li, H.: Inductive expression of the NOD1 signalling pathway in chickens infected with Salmonella pullorum, Br. Poult. Sci., 17, 1–9, 2017.
Uchikawa, E., Lethier, M., Malet. H., Brunel, J., Gerlier, D., and Cusack, S.: Structural analysis of dsRNA binding to anti-viral pattern recognition receptors LGP2 and MDA5, Mol. Cell, 62, 586–602, 2016.
Wan, Q., Wang, L., Su, J., Yang, C., Peng, L., and Chen, L.: Genetic structure, polymorphism identification of LGP2 gene and their relationship with the resistance/susceptibility to GCRV in grass carp, Ctenopharyngodon idella, Gene, 521, 166–175, 2013.
Werling, D., Jann, O. C., Offord, V., Glass, E. J., and Coffey, T. J.: Variation matters: TLR structure and species-specific pathogen recognition, Trends Immunol., 30, 124–130, 2009.
Zhu, Z., Zhang, X., Wang, G., and Zheng, H.: The laboratory of genetics and physiology 2: Emerging insights into the controversial functions of this RIG-I-like receptor, BioMed Res. Int., 2014, 960190, https://doi.org/10.1155/2014/960190, 2014.