the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The novel T755C mutation in BMP15 is associated with the litter size of Iranian Afshari, Ghezel, and Shal breeds
Hamid-Reza Amini
Amir Ajaki
Majid Farahi
Mitra Heidari
Ahmad Pirali
Mohsen Forouzanfar
Shahin Eghbalsaied
The present study was conducted to determine the molecular genetic variation in bone morphogenetic protein 15 (BMP15) of some selected Iranian sheep breeds and aims to provide relevant genetic information for twin-birth lambing. Therefore, a total of 44 rams and 213 ewes with single-, twin-, and triplet-birth lambing were used from Shal, Ghezel, Afshari and Lori-Bakhtiari sheep breeds. Three primer pairs were used for amplification of exons 1 and 2 of the BMP15 gene. The sequencing result of distinctive single-strand conformation polymorphism (SSCP) patterns showed segregation of the T755C mutation and the ensuing non-conservative substitution of L252P in the BMP15 propeptide. All three ewes with triplet-birth lambing and two sterile ewes were heterozygotic for this mutation. Association analysis revealed that this mutation was significantly related to the ewe's litter size, so that ewes with the CT genotype had 0.24 and 0.30 more lambs than those with the CC (p < 0.05) and TT (p < 0.01) genotypes, respectively. The mutation effect on the litter size was incomplete dominance in Afshari breed and over-dominance in Shal and Ghezel breeds. In conclusion, the findings in this experiment suggested synergistic effects of the T755C mutation and other unknown mutations in other effective genes which cause infertility and higher litter size in two different ways.
- Article
(2951 KB) -
Supplement
(811 KB) - BibTeX
- EndNote
The efficiency of sheep production is conditioned by fertility. Therefore, detection of genetic variation in major genes which affect prolificacy traits is an important prerequisite to improve the quantity, quality, and diversity of production in the livestock industry (Notter, 1999). Antral follicle count, ovulation rate, and litter size can be regulated by a set of major genes that mainly belong to the bone morphogenetic protein (BMP) signalling pathway (Hanrahan et al., 2004; Davis, 2005; Fabre et al., 2006; Pelosi et al., 2015). These fecundity genes include bone morphogenetic protein receptor 1β (BMPR1b or FecB), growth and differentiation factor 9 (GDF9 or FecG), and bone morphogenetic protein (BMP15 or FecX), which encode the most important growth factors involved in folliculogenesis (Mermillod et al., 2008). BMP15 is involved in the development of ovarian follicles and is a key regulator of some processes in the granulosa cells, including cellular proliferation, apoptosis prevention, and steroidogenesis (Juengel and McNatty, 2005).
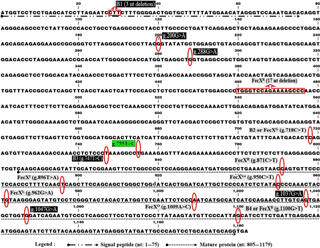
Figure 1The location and effect of various mutations in ovine BMP15 mRNA (ENSOARG00000009372) on ewe prolificacy. Mutations in the gray box are the effect on the ewe litter size. Mutations in the black box have not been documented to have an effect on the ewe litter size. The T755C mutation, the green box, is the novel mutation that has been detected in Iranian sheep breeds of this study.
In sheep, the BMP15 gene is located on chromosome X and encompasses two exons, which are separated via a 5.4 kb intron. This gene has a 1179 base pair open reading frame (ORF) encoding an immature polypeptide with 393 amino acid residues and an active mature peptide that is 125 amino acids long (Galloway et al., 2000). As reviewed in Fig. 1, 15 mutations have heretofore been identified in the sheep BMP15 gene. Among them, six mutations of FecXI, FecXH, FecXB, FecXG, FecXL, and FecXR increase ovulation rate and litter size in heterozygous animals, while completely sterilizing in homozygous mutants (Galloway et al., 2000; Hanrahan et al., 2004; Bodin et al., 2007; Martinez-Royo et al., 2008; Monteagudo et al., 2009). In contrast, homozygous ewes for two mutations of FecXGr and FecX0 are unexpectedly hyper-prolific and hence regarded as striking variants in the sheep BMP15 gene (Demars et al., 2013). The other six mutations, including the CTT deletion in signal peptide (B1), g.200G > A, g.288G > A, and g.747T > C (B3) in the proprotein region, as well as g.1037G > A and g.1047G > A in the mature protein, have been reported as ineffective variants. With the recent progress in the genetics field, there is considerable evidence that the presence of silent mutations (such as B3, g.288G > A, and g.1047G > A) can alter the protein functionality (Parmley and Hurst, 2007; Sauna et al., 2007; Hunt et al., 2014) and the phenotype variation through splicing accuracy, translation loyalty, mRNA structure, and miRNA binding (Chamary and Hurst, 2005; Sauna and Kimchi-Sarfaty, 2011; Cuevas et al., 2011). However, there is no available data to attest to or controvert this approach for litter size trait. Conversely, although numerous mutations have been detected in major genes relevant to prolificacy of various sheep breeds around the world, the Lleyn breed and many other highly prolific ewe breeds with triplet-birth records, did not carry these known significant mutations (Mullen et al., 2013). Therefore, screening highly prolific ewes for the known genes and other candidate genes is essential to fulfill our knowledge about these intra- and interbreed genetic variations.
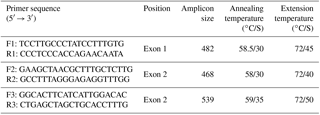
Table 2Primers and PCR conditions utilized for amplification of exon 1 and exon 2 of the sheep BMP15 gene.
Over the last decade, several studies have been conducted on screening mutations in the BMP15 gene of Iranian sheep breeds (Barzegari et al., 2010; Hafezian, 2011; Javanmard et al., 2011; Eghbalsaied et al., 2012; Zamani et al., 2015; Doran et al., 2016; Ahmadi et al., 2016;Nadri et al., 2016; Abdoli et al., 2017). Although novel mutations have been detected in BMP15 in some Iranian sheep breeds (Asghari et al., 2009; Javanmard et al., 2011; Zamani et al., 2015; Ahmadi et al., 2016; Nadri et al., 2016; Abdoli et al., 2017), these mutations have either been non-significant or had a partial effect on litter size in these breeds. Therefore, responsible mutations in BMP15 have yet to be discovered in these breeds. Thus, the aim of the present study was to screen the BMP15 gene in a group of infertile, twin-birth, and triplet-birth ewes and to evaluate the mutation effect on the litter size of Iranian Shal, Afshari, Ghezel, and Lori-Bakhtiari breeds. These breeds are the most prominent native sheep in northern, central, and southern parts of the Zagros mountain chain and have a large carcass and fat-tail size (Eskandarinasab et al., 2010; Vatankhah et al., 2016).
2.1 Animals and sample collection
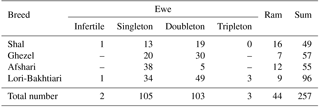
All the following procedures which were carried out on animals were approved by the Animal Welfare Committee of Isfahan (Khorasgan) branch of the Islamic Azad University. The total genotyped samples were comprised of 257 sheep with 49 Shal, 57 Ghezel, 55 Afshari, and 96 Lori-Bakhtiari sheep (Table 1). In this study, blood samples of Shal, Ghezel, Afshari, and Lori-Bakhtiari sheep breeds were obtained from the Bouinzahra region (Qazvin Province, situated at 36∘16′ N and 50∘00′ E); Miyandoaab Research Center and the Simineh (Bukan) region (West Azerbaijan Province, located at 36.92∘ N and 46.16∘ E); Khatoon Abad region (Isfahan Province, located at 33.27∘ N and 52.36∘ E); and the three regions of Shahr-e Kord, Lordegan, and Farsan (Chaharmahal and Bakhtiari Province, situated at 32.32∘ N and 50.85∘ E), respectively.
2.2 DNA extraction and PCR amplification
Genomic DNA was extracted from the whole blood by the phenol–chloroform method (Sambrook and Russell, 2006). Final DNA pellet was dissolved in the double distilled water and was stored at −20 ∘C for further experiments. Exon 1 and exon 2 of BMP15 were amplified by three specific primers which were designed using the Primer3 software from the NCBI website (Table 2). Polymerase chain reaction (PCR) was conducted in 25 µL total volume containing approximately 50–100 ng of genomic DNA, 2.5 mM of MgCl2, 10 × PCR buffer (500 mM of KCl and Tris HCl (pH 8.4)), 10 µM of forward–reverse primer, 0.2 mM of dNTPs, and 1 IU Smart Taq DNA polymerase (catalogue number: TA8108C, SinaClon, Iran). The PCR amplifications were carried out in a thermal cycler (Bio-Rad, USA) by using a specific annealing temperature (Table 2).
2.3 SSCP analysis
The genotypic patterns of PCR products were determined using the single-strand conformation polymorphism (SSCP) method with slight modifications (Orita et al., 1989). Briefly, a constant volume of 2 µL PCR products of each sample was mixed with 8 µL of gel loading dye solution (98 % formamide, 10 mM EDTA, 0.025 % bromophenol blue, and 0.025 % xylene cyanol). The mixture was vortexed and denatured at 95 ∘C for 10 min and rapidly chilled on ice and loaded on 10 % polyacrylamide gels (acrylamide : bisacrylamide = 37.5 : 1). Electrophoresis was carried out at 250 V for 24 h at 4 ∘C in 1 × TBE buffer. Then, the DNA bands were visualized using silver staining (Bassam et al., 1991). After the SSCP analysis, three samples from each distinctive pattern were sequenced using both the forward and reverse primers (Bioneer, Korea). The sequences were analysed using the ClustalW subprogram of BioEdit software to execute the multiple sequence alignments (Hall et al., 2011).
2.4 Sequence and statistical analysis
The genotype and allele frequency, as well as Hardy–Weinberg equilibrium test, were estimated using the GenAlEx 6.5 program (Peakall and Smouse, 2006). Polymorphism information content (PIC) values were computed using HET software version 1.8 (Ott, 2001). The GENMOD procedure in the SAS software was implemented to analyse the association between genetic variation in the BMP15 gene and the litter size records (Institute, 1985). Also, mean comparison between different genotypes was carried out using the LSM (least-square mean) option. A P value ≤ 0.05 was considered as the statistically significant level.
3.1 PCR–SSCP and sequencing
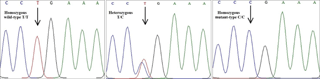
The coding sequence for the BMP15 gene of four Iranian sheep breeds was amplified using three primer pairs, and the amplified fragments were used for PCR–SSCP. After optimization of the SSCP procedure, observation of different banding patterns suggested the presence of variant sequences in the amplified regions. For validation of the attained results from the PCR–SSCP, sequencing of distinct SSCP patterns was also carried out (Bioneer, Korea). Based on the sequencing results of the PCR and their subsequent alignment with the reference sequence of the BMP15 gene on the NCBI website (accession number NC_019484), a T755C mutation was detected. This mutation has recently been detected in exon 2 of BMP15 using next-generation sequencing data in Iranian Ovis aries (accession number rs55628000) (Fig. 2). Multiple alignments of the flanking sequence of this single-nucleotide polymorphism (SNP) with other mammals revealed that this mutation takes place in a conserved region that corresponds to the pro-region of the BMP15 protein.
3.2 Allelic and genotypic frequencies
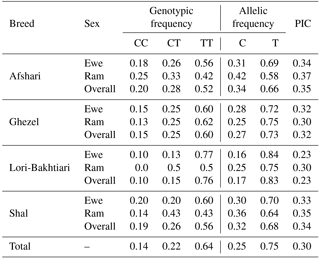
Genotypes and allele frequencies of c.755T > C SNPs in BMP15 gene of the Shal, Ghezel, Lori-Bakhtiari and Afshari sheep breeds are presented in Table 3. Three genotypic patterns of this SNP were detected in all four of the breeds. This SNP displayed a higher frequency of T than the C allele (25 % frequency in the whole population), and the TT genotype predominated in the whole population (average = 0.64). The allelic frequency of the T755C mutation ranged from 0.25 (Lori-Bakhtiari and Ghezel) to 0.42 (Afshari) in rams and 0.16 (Lori-Bakhtiari) to 0.31 (Afshari) in ewes. None of the studied breeds were in the Hardy–Weinberg equilibrium (p < 0.01). The genetic diversity for c.755T > C locus in the BMP15 gene was at the medium level in all populations, except for the Lori-Bakhtiari breed.
3.3 Association analysis
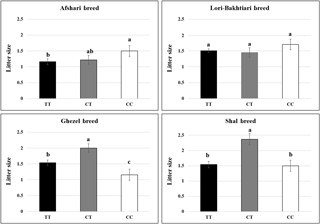
The LSMs of litter size for different genotypes of the BMP15 gene are shown in Fig. 3. The association analysis showed that this mutation had a significant effect on the ewe's litter size (P value = 0.009). Considering the genotype effect on lambing size in different breeds indicated that the CT genotypes had a higher litter size than the TT (0.5 and 0.8 lambs) and CC groups (0.7 and 0.9 lambs) in the Shal and Ghezel breeds, respectively. However, the homozygous mutant genotype was associated with higher lambing compared to the heterozygous animals of the Afshari breed (P value = 0.022). Also, ewes with sterile and tripleton phenotypes were heterozygous for this mutation.
It has been well documented that heterozygote carriers for BMP15 mutations display an increased ovulation rate and litter size compared to the wild-type ewes (Galloway et al., 2000; Hanrahan et al., 2004; Bodin et al., 2007; Martinez-Royo et al., 2008; Monteagudo et al., 2009). The majority of these mutations were located in the propeptide region of BMP15 or GDF9 (Di Pasquale et al., 2004; Inagaki and Shimasaki, 2010; Abdoli et al., 2016). The propeptide region of the protein is one of the most important attributes affecting the post-translational processing of the transforming growth factor beta (TGF β) superfamily (Liao et al., 2003; Shimasaki et al., 2004; Di Pasquale et al., 2004).
In the current study, we screened BMP15 in a group of ewes with single-, twin-, and triplet-birth lambing and a sample of rams as flock sires in the four main breeds Lori-Bakhtiari, Shal, Ghezel, and Afshari. One SNP was detected with PCR–SSCP and verified by sequencing of the amplicons. Except for the GWAS data, this is the first report on the presence and importance of this SNP (T755C) for ewe's litter size (rs55628000). From 20 genotyped samples of Iranian Ovis aries sheep, the frequency of 5 % was reported for the T755C mutation using next-generation whole-genome sequencing. However, the current paper is the first report which provides the population and association study on this mutation. The frequency of this mutation varied from 0.17 to 0.34 in Lori-Bakhtiari and Afshari breeds, respectively. Lack of Hardy–Weinberg equilibrium in this study implied that natural or artificial selection favoured the mutated allele in all four breeds. The majority of sheep breeds in Iran are kept under the traditional animal husbandry system in rustic and nomadic production systems. Thus, this unequal distribution indicates that the T755C mutation can be associated with fitness traits which form the main practical index in the traditional breeding systems of sheep species. Also, the PIC index indicated the presence of a moderate genetic diversity for this mutation, which makes it plausible for being included in the breeding programs (Botstein et al., 1980). In our study, the ewes with the CT genotype had more lambs than those with homozygous genotypes. This indicates that the mutation has a dominance effect on the ewe litter size. It might be noteworthy that the infertile ewes were also heterozygous for this mutation. Conversely, all ewes with triplet-birth phenotypes were heterozygous carriers for this locus. These unexpected results strongly suggested synergistic effects of the T755C mutation with other mutation(s), which may induce over-dominance and sterility when heterozygous and homozygous, respectively. Therefore, other mutations in the non-sequenced region of BMP15 or other above-mentioned genes may act through a nonadditive effect with the T755C mutation and induce infertility or higher prolificacy. Similar to some of the FecX variants that have been detected, the prolific ewes with the heterozygote T755C genotype had an increased litter size, while no sterility phenotype was detected in homozygous animals. However, no significant differences between heterozygous and homozygous mutant types were observed in the Lori-Bakhtiari or Afshari sheep breeds. The reason for these interbreed differences can be related to the difference in the genetic background of these breeds (Eghbalsaied et al., 2017). Our recent study on GDF9 mutations confirmed the high variation in the frequency of G1 and G4 SNPs in these four evaluated breeds (Eghbalsaied et al., 2017). Sequencing of GDF9 did not show any significant mutation in these infertile and triplet-birth ewes (Eghbalsaied et al., 2017) as well as ewes with high antral follicle count (Eghbalsaied et al., 2012). This may suggest the presence of the T755C mutation with a different set of SNPs in other candidate genes, such as AMPK (Foroughinia et al., 2017), Cyp19 (Foroughinia et al., 2017), or leptin (Juengel et al., 2015) in different sheep breeds.
The results of this study documented the segregation of the T755C mutation, T755C, in BMP15 in the Shal, Ghezel, Afshari, and Lori-Bakhtiari sheep breeds. This mutation causes a non-conservative substitution of L252P in the BMP15 propeptide. Association analysis revealed the significance of this mutation on the litter size, so that the ewes with the CT genotype had more lambs than those with either CC or TT genotypes. Also, all ewes with triplet-birth lambing and sterile ewes were heterozygous for this mutation. The results suggested a synergistic effect of the T755C SNP and other unknown mutations in other important genes which cause infertility and higher litter size in two different ways.
The data used in this study are available in the Supplement.
The supplement related to this article is available online at: https://doi.org/10.5194/aab-61-153-2018-supplement.
SE conceived and designed the experiments. HRA, AA, MF, AP, MH, MF, and SE conducted the experiments. HRA and SE analysed the data. HRA and SE wrote the paper.
The authors have no conflict of interest to declare.
The authors would like to thank Alan McNeilly for his invaluable
collaboration in conducting this project and Mehdi Pourmasoum for his
assistance in the blood collection. This project was supported by the Isfahan
(Khorasgan) branch of the Islamic Azad University, Iran, through grant number
51755920814006.
Edited by: Steffen
Maak
Reviewed by: Pouya Zamani and Hamed Amirpour Najafabadi
Abdoli, R., Zamani, P., Mirhoseini, S., Ghavi Hossein-Zadeh, N., and Nadri, S.: A review on prolificacy genes in sheep, Reproduction in Domestic Animals, 51, 631–637, 2016.
Abdoli, R., Mirhoseini, S. Z., Hossein-Zadeh, N. G., and Zamani, P.: Screening for causative mutations of major prolificacy genes in Iranian fat-tailed sheep, Int. J. Fertil. Steril., 12, 51–55, 2017.
Ahmadi, A., Afraz, F., Talebi, R., Farahavar, A., and Vahidi, S.: Investigation of GDF9 and BMP15 polymorphisms in Mehraban sheep to find the missenses as impact on protein, Iranian Journal of Applied Animal Science, 6, 863–872, 2016.
Asghari, S. R., Irajeyan, G. H., and Buesagh, H.: Genetic polymorphism FecB and BMP15 genes and its association with litter size in Sangsari sheep breed of Iran, J. Anim. Vet. Adv., 8, 1025–1031, 2009.
Barzegari, A., Atashpaz, S., Ghabili, K., Nemati, Z., Rustaei, M., and Azarbaijani, R.: Polymorphisms in GDF9 and BMP15 associated with fertility and ovulation rate in Moghani and Ghezel sheep in Iran, Reprod. Domest. Anim., 45, 666–669, 2010.
Bassam, B. J., Caetano-Anollés, G., and Gresshoff, P. M.: Fast and sensitive silver staining of DNA in polyacrylamide gels, Anal. Biochem., 196, 80–83, 1991.
Bodin, L., Di Pasquale, E., Fabre, S., Bontoux, M., Monget, P., Persani, L., and Mulsant, P.: A novel mutation in the bone morphogenetic protein 15 gene causing defective protein secretion is associated with both increased ovulation rate and sterility in Lacaune sheep, Endocrinology, 148, 393–400, 2007.
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W.: Construction of a genetic linkage map in man using restriction fragment length polymorphisms, Am J. Hum. Genet., 32, 314–331, 1980.
Chamary, J. and Hurst, L. D.: Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals, Genome. Biol., 6, 1–12, 2005.
Cuevas, J. M., Domingo-Calap, P., and Sanjuán, R.: The fitness effects of synonymous mutations in DNA and RNA viruses, Mol. Biol. Evol., 29, 17–20, 2011.
Davis, G. H.: Major genes affecting ovulation rate in sheep, Genet. Sel. Evol., 37, S11-S23, 2005.
Demars, J., Fabre, S., Sarry, J., Rossetti, R., Gilbert, H., Persani, L., Tosser-Klopp, G., Mulsant, P., Nowak, Z., and Drobik, W.: Genome-wide association studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy phenotype in sheep, PLoS Genet, 9, e1003482, https://doi.org/10.1371/journal.pgen.1003482, 2013.
Di Pasquale, E., Beck-Peccoz, P., and Persani, L.: Hypergonadotropic ovarian failure associated with an inherited mutation of human bone morphogenetic protein-15 (BMP15) gene, Am. J. Hum. Genet., 75, 106–111, 2004.
Doran, T. J., Cooper, C. A., Jenkins, K. A., and Tizard, M. L.: Advances in genetic engineering of the avian genome:“Realising the promise”, Transgenic Res., 25, 307–319, 2016.
Eghbalsaied, S., Ghaedi, K., Shahmoradi, S., Pirestani, A., Amini, H., Saiedi, T., Nicol, L., and McNeilly, A.: Presence of SNPs in GDF9 mRNA of Iranian Afshari sheep, International journal of fertility & sterility, 5, 225–230, 2012.
Eghbalsaied, S., Khorasgani, F. R., Amini, H.-R., Farahi, M., Davari, M., Pirali, A., Pourali, S., Vatankhah, M., Rostami, M., and Atashi, H.: Variant GDF9 mRNA is likely not the main cause of larger litter size in Iranian Lori-Bakhtyari, Shal, Ghezel, and Afshari sheep breeds, Arch. Anim. Breed., 60, 119–129, https://doi.org/10.5194/aab-60-119-2017, 2017.
Eskandarinasab, M., Ghafouri-Kesbi, F., and Abbasi, M.: Different models for evaluation of growth traits and Kleiber ratio in an experimental flock of Iranian fat-tailed Afshari sheep, J. Anim. Breed. Genet., 127, 26–33, 2010.
Fabre, S., Pierre, A., Mulsant, P., Bodin, L., Di Pasquale, E., Persani, L., Monget, P., and Monniaux, D.: Regulation of ovulation rate in mammals: contribution of sheep genetic models, Reprod. Biol. Endocrin., 4, 20, https://doi.org/10.1186/1477-7827-4-20, 2006.
Foroughinia, G., Fazileh, A., and Eghbalsaied, S.: Expression of genes involved in BMP and estrogen signaling and AMPK production can be important factors affecting total number of antral follicles in ewes, Theriogenology, 91, 36–43, 2017.
Galloway, S. M., McNatty, K. P., Cambridge, L. M., Laitinen, M. P., Juengel, J. L., Jokiranta, T. S., McLaren, R. J., Luiro, K., Dodds, K. G., and Montgomery, G. W.: Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner, Nat. Genet., 25, 279–283, 2000.
Hafezian, S. H.: Genetic polymorphism BMP15 and GDF9 genes in Sangsari sheep of Iran, International Journal of Genetics and Molecular Biology, 3, 31–34, 2011.
Hall, T., Biosciences, I., and Carlsbad, C.: BioEdit: an important software for molecular biology, GERF Bull Biosci., 2, 60–61, 2011.
Hanrahan, J. P., Gregan, S. M., Mulsant, P., Mullen, M., Davis, G. H., Powell, R., and Galloway, S. M.: Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries), Biol. Reprod., 70, 900–909, 2004.
Hunt, R. C., Simhadri, V. L., Iandoli, M., Sauna, Z. E., and Kimchi-Sarfaty, C.: Exposing synonymous mutations, Trends Genet., 30, 308–321, 2014.
Inagaki, K. and Shimasaki, S.: Impaired production of BMP-15 and GDF-9 mature proteins derived from proproteins with mutations in the proregion, Mol. Cell. Endocrinol., 328, 1–7, 2010.
Institute, S.: SAS user's guide: statistics, Sas Institute, USA, 1985.
Javanmard, A., Azadzadeh, N., and Esmailizadeh, A. K.: Mutations in bone morphogenetic protein 15 and growth differentiation factor 9 genes are associated with increased litter size in fat-tailed sheep breeds, Vet. Res. Commun., 35, 157–167, 2011.
Juengel, J. and McNatty, K.: The role of proteins of the transforming growth factor-β superfamily in the intraovarian regulation of follicular development, Hum. Reprod. Update, 11, 144–161, 2005.
Juengel, J. L., French, M. C., O'Connell, A. R., Edwards, S. J., Haldar, A., Brauning, R., Farquhar, P. A., Dodds, K. G., Galloway, S. M., and Johnstone, P. D.: Mutations in the leptin receptor gene associated with delayed onset of puberty are also associated with decreased ovulation and lambing rates in prolific Davisdale sheep, Reproduction, Fertility and Development, 28, 1318–1325, 2015.
Liao, W. X., Moore, R. K., Otsuka, F., and Shimasaki, S.: Effect of intracellular interactions on the processing and secretion of bone morphogenetic protein-15 (BMP-15) and growth and differentiation factor-9 Implication of the aberrant ovarian phenotype of BMP-15 mutant sheep, J. Biol. Chem., 278, 3713–3719, 2003.
Martinez-Royo, A., Jurado, J., Smulders, J., Marti, J., Alabart, J., Roche, A., Fantova, E., Bodin, L., Mulsant, P., and Serrano, M.: A deletion in the bone morphogenetic protein 15 gene causes sterility and increased prolificacy in Rasa Aragonesa sheep, Anim. Genet., 39, 294–297, 2008.
Mermillod, P., Dalbiès-Tran, R., Uzbekova, S., Thelie, A., Traverso, J. M., Perreau, C., Papillier, P., and Monget, P.: Factors affecting oocyte quality: who is driving the follicle?, Reprod. Domest. Anim., 43, 393–400, 2008.
Monteagudo, L. V., Ponz, R., Tejedor, M. T., Laviña, A., and Sierra, I.: A 17bp deletion in the Bone Morphogenetic Protein 15 (BMP15) gene is associated to increased prolificacy in the Rasa Aragonesa sheep breed, Anim. Reprod. Sci., 110, 139–146, 2009.
Mullen, M. P., Hanrahan, J. P., Howard, D. J., and Powell, R.: Investigation of Prolific Sheep from UK and Ireland for Evidence on Origin of the Mutations in BMP15 (FecXG, FecXB) and GDF9 (FecGH) in Belclare and Cambridge Sheep, PloS one, 8, e53172, https://doi.org/10.1371/journal.pone.0053172, 2013.
Nadri, S., Zamani, P., and Ahmadi, A.: Novel mutation in Exon 1 of the BMP15 gene and its association with reproduction traits in sheep, Anim. Biotechnol., 27, 256–261, 2016.
Notter, D. R.: The importance of genetic diversity in livestock populations of the future, J. Anim. Sci., 77, 61–69, 1999.
Orita, M., Iwahana, H., Kanazawa, H., Hayashi, K., and Sekiya, T.: Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms, P. Natl. Acad. Sci. USA, 86, 2766–2770, 1989.
Ott, J.: Program HET Version 1.8. Utility programs for analysis of genetic linkage, Rockefeller University, NY, USA, available at: http://linkage.rockefeller.edu/software/utilities, 2001.
Parmley, J. L. and Hurst, L. D.: How do synonymous mutations affect fitness?, Bioessays, 29, 515–519, 2007.
Peakall, R. and Smouse, P. E.: GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research, Mol. Ecol. Notes, 6, 288–295, 2006.
Pelosi, E., Forabosco, A., and Schlessinger, D.: Genetics of the ovarian reserve, Frontiers in genetics, 6, 308, https://doi.org/10.3389/fgene.2015.00308, 2015.
Sambrook, J. and Russell, D. W.: Purification of nucleic acids by extraction with phenol: chloroform, Cold Spring Harbor Protocols, pdb. prot4455, https://doi.org/10.1101/pdb.prot4455, 2006.
Sauna, Z. E. and Kimchi-Sarfaty, C.: Understanding the contribution of synonymous mutations to human disease, Nat. Rev. Genet., 12, 683–691, 2011.
Sauna, Z. E., Kimchi-Sarfaty, C., Ambudkar, S. V., and Gottesman, M. M.: Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer, Cancer Res., 67, 9609–9612, 2007.
Shimasaki, S., Moore, R. K., Otsuka, F., and Erickson, G. F.: The bone morphogenetic protein system in mammalian reproduction, Endocr. Rev., 25, 72–101, 2004.
Vatankhah, M., Talebi, M., and Blair, H.: Genetic analysis of Lori-Bakhtiari lamb survival rate up to yearling age for autosomal and sex-linked, Small Ruminant Res., 136, 121–126, 2016.
Zamani, P., Nadri, S., Saffaripour, R., Ahmadi, A., Dashti, F., and Abdoli, R.: A new mutation in exon 2 of the bone morphogenetic protein 15 gene is associated with increase in prolificacy of Mehraban and Lori sheep, Trop. Anim. Health Pro., 47, 855–860, 2015.